Home »
Cloud Computing
Cloud Application: Protein Structure Prediction
Protein Structure Prediction: In this tutorial, we will learn how protein structure prediction can be carried out using cloud computing, why we need Protein, and the most common instruments for the prediction of protein structures.
By IncludeHelp Last updated : June 02, 2023
What is Protein Structure Prediction in Cloud Computing?
The prediction of the protein structure is the inference from its amino acid sequence of the three-dimensional structure of a protein, that is, the prediction of its secondary and tertiary structure from the primary structure.
Proteins are large amino acid-based molecules that our bodies and the cells in our bodies need to function correctly. Significant quantities of protein are present in our muscles, skin, bones, and many other areas of the body. The prediction of protein structure is the best example in the field of science that makes use of cloud applications for computing and storage. First, the primary protein structures are formed, and then the secondary, tertiary, and quaternary structures are predicted from the primary structure.
Predictions of protein structures are carried out in this manner. The prediction of protein structures also makes use of numerous other technologies, such as artificial neural networks, artificial intelligence, machine learning, and probabilistic approaches, and is also of great significance in fields such as theoretical chemistry and bioinformatics.
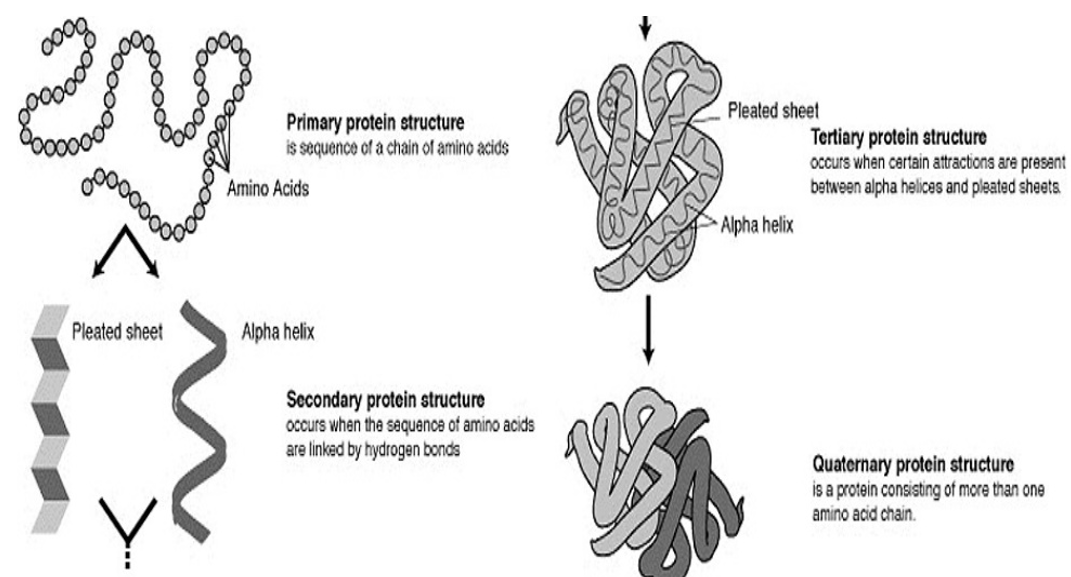
Fig: Composition of the Protein
Why do we need Protein?
Applications or s/w that require high computing capabilities and have large data sets will result in high I/O operations. The prediction of the protein structure will enable the production of new drugs by medical scientists.
System administrators should take advantage of a range of resources to monitor and manage the technology deployed. This can be a public cloud accessible via the Internet to everyone, or a private cloud created by a group of access-restricted nodes.
These approaches can turn the problem in such a way that they can be divided into three phases: initialization, classification, and a final step. Researchers can not only easily deploy their prediction applications in a distributed environment using the Grid middleware, but also track and control the execution in the distributed environment.
Most Common Instruments for Prediction Of Protein Structures
1) CASP
CASP (Critical Assessment of Structure Prediction) is a community-wide experiment in the simulation of protein structure from the amino acid sequence to define and advance the state of the art. CASP tests many aspects of modeling, including the precision of topologies of proteins, coordinates of atoms, and assemblies of multi-proteins. The experiment also explores the degree to which models can address biological-interest questions and how various types of experimental data with sparse or low resolution can boost the accuracy of the model.
2) CAMEO
Continuous Automated Model Evaluation (CAMEO) is a community-wide project that is fully automated to continuously evaluate the accuracy and reliability of protein structure prediction servers.
Advertisement
Advertisement